→ Projects for internships
Students interested in developing computational models to understand root development, please see below the list of available projects in the lab and feel free to
contact me to share more details.
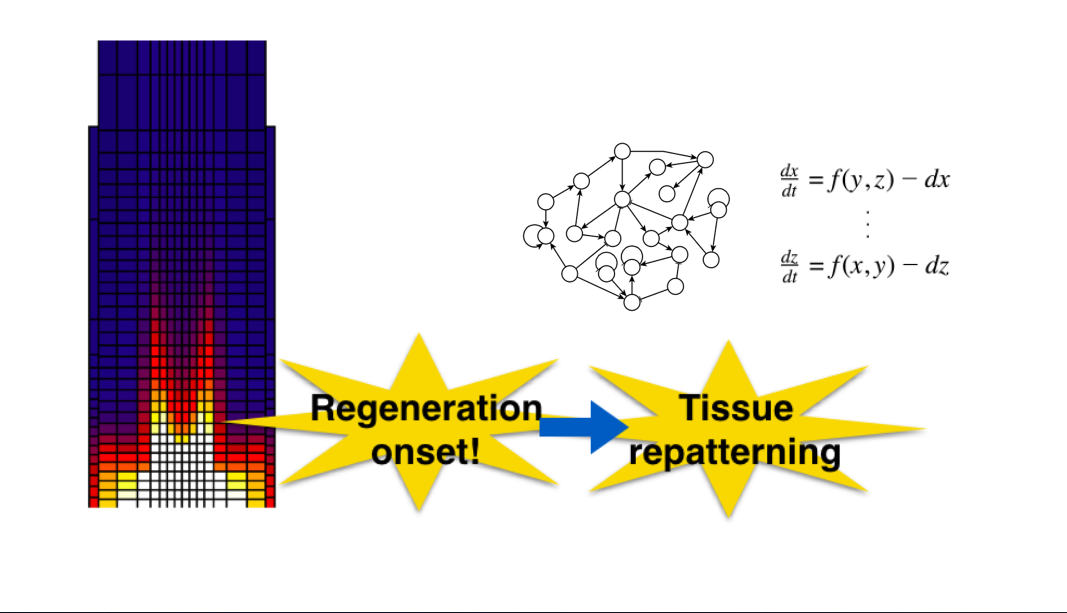
Regulatory networks controlling stem cell niche patterning in uncut and cut roots: here you will explore the genetic-hormonal regulatory network that underlies the establishment of a root stem cell niche de novo. For this you will use a multi-scale model of the root tip, where you will simulate root cuts and recover the ordered cell fate changes observed during the regeneration process.
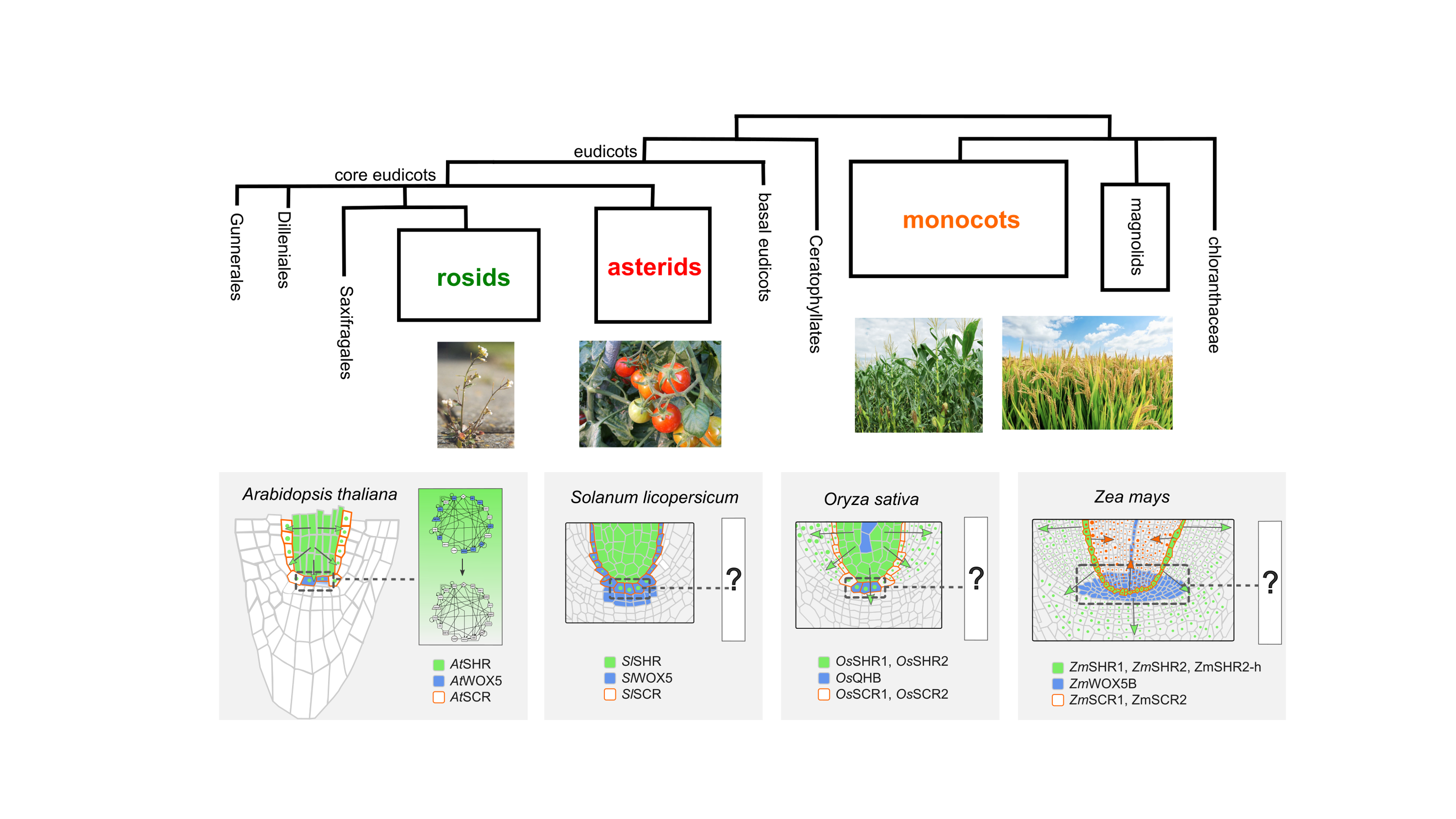
 Modelling Root Stemness: From A Simple Plant To Crops: here you will use computational modelling and bioinformatics to explore how the mechanism regulating stem cell maintenance work in different plant species.
Modelling Root Stemness: From A Simple Plant To Crops: here you will use computational modelling and bioinformatics to explore how the mechanism regulating stem cell maintenance work in different plant species.
 Agent based models of the intracellular transport of key root patterning regulators: here you will study the strategies that allow proteins to rapidly reach a target (plasmodesmata) without prior information. This model is centered on mobile transcription factors that play a key role in root patterning, and you will assess the impact of different transport strategies on the organization of the root stem cell niche.
Agent based models of the intracellular transport of key root patterning regulators: here you will study the strategies that allow proteins to rapidly reach a target (plasmodesmata) without prior information. This model is centered on mobile transcription factors that play a key role in root patterning, and you will assess the impact of different transport strategies on the organization of the root stem cell niche.