The Virtual Laboratory Team

|
Bram van Dijk, withSultan A. Nazir (PhD candidate) |
| Home | Education | Google Scholar | Personal website |
Research interests
It captivates me how biological complexity is often governed by very simple
rules. I focus my scientific energy on simulating complex microbial ecosystems and identifying key principles of their evolution. These models contain many interacting players, such as genes, mobile genetic elements, microbes, and their hosts like plants and animals. I believe that these computational models are integral to understanding the complexity of nature.
Microbial ecology and evolution: a multi-level perspective
Ever since I first learned about the astonishing diversity of microbial communities, I have been unable to stop thinking about these mind-boggling systems. Although our grasp on microbial communities is still lacking, they are an indispensable element of life on Earth. Microbial communities are responsible for completing nutrient cycles in terrestrial, and aquatic ecosystems, and moreover, form intricate symbiotic relationships with larger organisms such as plants and animals. Furthermore, microbial community function has more direct implications for human well-being in terms of the production of antibiotics, gut and skin health, and its relationship to the human immune system.
While it is certainly useful to study complex communities by reducing them to fewer players — e.g. by cultivating small communities in the lab or by using simple mathematical models — such simplification may reduce our ability to understand the processes that are inherent to the complexity itself. In addition to simplification, I argue that embracing the astonishing complexity of microbial communities in our models is a surprisingly productive starting point. This entails implementing models of microbial ecology and evolution with multiple levels of organisation, multiple (potentially conflicting) selection pressures, non-trivial genotype-to-phenotype mapping, and sufficient potential to form complex interactions. Although investigating these models is a major challenge, I believe that these systems are essential to cross-pollinate with experimentalists and bioinformaticians, allowing us to truly get a handle on the unfathomable complexity of microbial communities.
To see a video of what such a multi-level system of ecology and evolution may look like, see the video below:
Co-evolution of mobile genetic elements, their hosts, and 'superhosts'
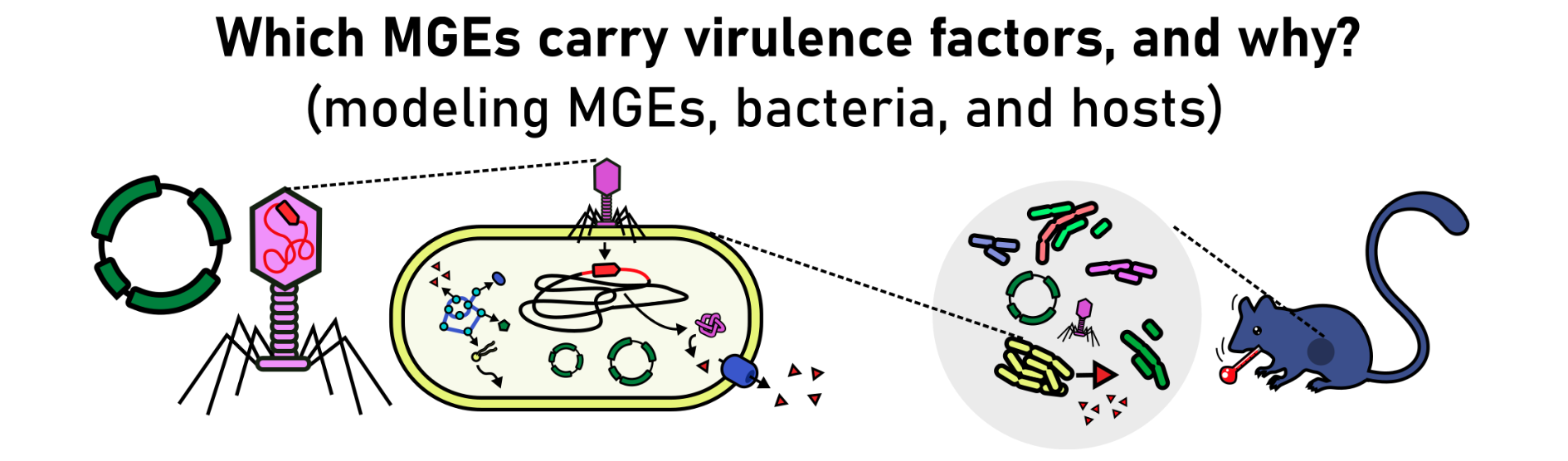
Microbial evolution is driven by rapid changes in gene content mediated by horizontal gene transfer (HGT). These rapid changes are promoted by mobile genetic elements (MGEs), entities that evolved to persist and replicate by moving DNA within and between microbial cells. MGEs are often considered to be on a continuum between parasitism and mutualism, sometimes benefitting their host, and sometimes merely replicating for their own gain. The precise relationships between MGEs and their hosts are complex, ever-changing, and highly dependent upon ecological context. In our team, we build multi-level models of microbial evolution, for example, to understand how MGEs co-evolve with microbial populations and how these interactions affect the health of plants, animals, and ecosystems.
The goal of this project is to understand which types of MGEs are linked to the emergence of plant or animal pathogens, for example by carrying virulence factors as cargo, or by silencing genes that are recognized by the immune system. Vice versa, it will also be interesting to understand why other host-beneficial traits (such as antimicrobial resistance) are less often associated with those same mobile elements, which we suspect has to do with their lifestyle (parasitic vs. mutualistic).
Modelling horizontal gene transfer
Horizontal gene transfer (HGT) is the exchange of genes between two organisms without the necessity of a parent-offspring relationship. While virtually non-existent in complex organisms such as ourselves, HGT is very common for bacteria and other microorganisms. By picking up genes via HGT, bacteria can rapidly adapt to new ecological opportunities. However, HGT also risky. For example, by taking up genes from the environment, organisms expose themselves to genetic parasites called selfish genetic elements (SGEs), which simply try to replicate for their own gain.
Through in silico modelling (i.e. computer simulations of biological systems), I study how HGT affects the spread and maintenance of genes, and how this depends on the fitness effects or functions of these genes. With help of these models, I try to investigate when horizontal gene transfer can come to the rescue of a bacterial population, and when it is nothing less than a catastrophe.
Published insights from in silico modelling of HGT:
- The costly uptake of genes can be adaptive for bacteria, but only for genes that are slightly beneficial (https://doi.org/10.7554/eLife.56801)
- Transposable elements, self-replicating DNA sequences, can promote the evolution of smaller genomes (https://doi.org/10.1101/2021.05.29.446280)
- High levels of population diversity can be maintained when genes for different functions transfer at different rates (https://doi.org/10.1093/gbe/evv255)
Open questions:
- When do transposable elements become linked to genes with certain ecological functions, such as antimicrobial resistance?
- Can transposable elements persist by relying on host-encoded enzymes in order to jump? (e.g. as appears to be the case for RAYT/REPIN transposases)
- What are the long-term consequences of evolving through HGT (like prokaryotes) and evolving through gene duplications (like eukaryotes)?
Contact
Dr. B. (Bram) van Dijk PhD (he/they)
Hugo R. Kruytgebouw
Padualaan 8
Room N605
3584CH Utrecht
The Netherlands
b.vandijk@uu.nl


