




Kerstin Huhn, Ying Miao, Ulrike Zentgraf, Stefan Bieker,
Maren Potschin, Gabi Eggers-Schumacher, Carles Llorca (Merit-fellow),
Lena Riester (from left to right) on a tour to the “Nebelhöhle”
Our group is interested in the regulatory network involved in
senescence regulation. Developmental as well as dark-induced leaf
senescence is accompanied by a massive change in the transcriptome
clearly implying an important role of transcription factors in these
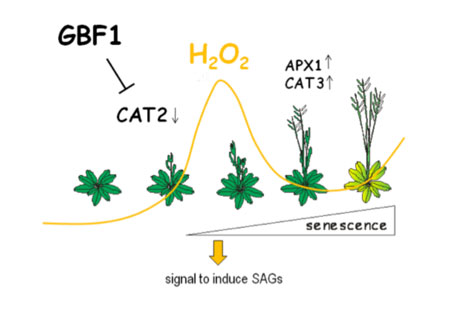
overlapping processes. We could show that G-box binding factor1 (GBF1),
belonging to the G-group bZIP transcription factors, is involved in the
induction of developmental leaf senescence since gbf1 mutant plants
exhibit a delayed senescence phenotype. GBF1 expression is not
transcriptionally up-regulated during leaf senescence and therefore
GBF1 is presumed to be post-transcriptionally regulated, possibly by
the formation of specific heterodimers with other bZIP factors. Another
possibility is that GBF1 is modified e.g. by specific kinases such as
SnRK1 or that it is regulated at the level of translation or its
intracellular localization in analogy with bZIP10. The aim of this
project will be to characterize the senescence-specific activation of
GBF1 and the crosstalk to low energy stress in more detail. Methodology
will include bimolecular fluorescence complementation assays (BiFC) and
FACS analyses, Yeast-two-hybrid, phosphorylation assays, site-directed
mutagenesis, senescence phenotyping and expression analyses,
ELISA-based DNA-binding assays, transient protoplast transformation,
intracellular hydrogen peroxide measurements.
 1)
Target genes and localization of GBF1/bZIP63 dimers.
1)
Target genes and localization of GBF1/bZIP63 dimers.
We will test the influence of GBF1/bZIP63 dimer formation on its
DNA-binding activity and on the expression of the reporter genes CAT2
and RUBISCO. We will follow up the intracellular localization of the
proteins and the complex over plant development. New target genes
should be isolated by ChIPSeq.
2) Influence of SnRK1 on GBF1/bZIP63 dimers.
bZIP63 is affected by SnRK1, which is related to starvation and the LES
in plants. Over-expressing a SnRK1 catalytic subunit increased the
expression of sugar-repressed and autophagy genes and showed a delayed
flowering and senescence phenotype (Wingler et al. (2009) J Ex. Bot 60
1063-1066). Therefore, we will analyze the interaction of bZIP63/GBF1
in the snrk1 mutant background and SnRK1 over-expressing lines and test
the expression of reporter genes and senescence induction. Conversely
dark-induced senescence will be compared between wild type plants and
gbf1 mutants. In addition, a direct interaction between SnRK1 and
GBF1/bZIP63 will be tested by means of Y2H, BiFC and Co-IP.
3) Identification of new interacting partners.
We will identify other bZIP protein interaction partners forming
heterodimers with GBF1 using a protoplast-based high through-put
system.







